xGen™ NGS Hybridization Capture
xGen NGS offers flexible hybridization capture solutions with quality, speed, and customer support for your research.
xGen NGS—made to deliver confidence.
Overview
Hybridization capture is a method of targeted next generation sequencing. Before hybridization capture is performed, DNA or RNA samples are converted into sequencing libraries. To create libraries, the sample is randomly sheared into smaller fragments by mechanical or enzymatic methods, and sequencing adapters are added. Depending on the library design, PCR amplification may be required.
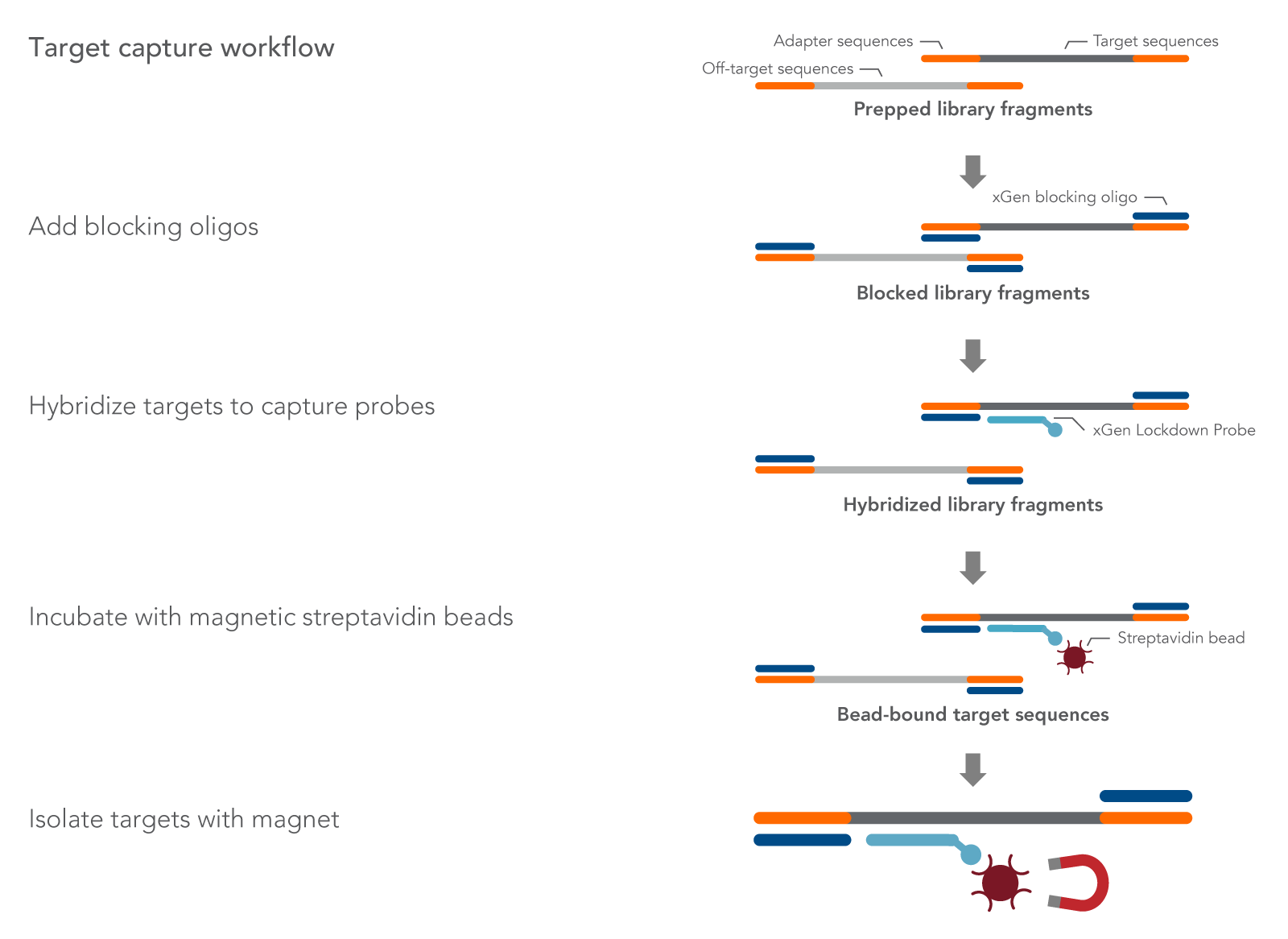
Regions of interest within the library are then captured using biotinylated oligonucleotide probes, sometimes referred to as baits (Figure 1). Biotinylated probes are designed to hybridize to regions of interest, such as exome, known genes related to cancer, or genes in a metabolic pathway. After hybridization of baits to the fragmented DNA or RNA, streptavidin is used to separate the probe and targeted fragment complex from other fragments that are not bound to probes. Probes can be arranged in different ways to sequence regions of interest. For example:
- Tiling probes allow regions that are sequenced to align end to end.
- Overlapping probes permit extra sequencing to occur at the ends of the targeted sequences to ensure that no part of the sequence is missed.
Probes can also be positioned to overcome challenging motifs such as repetitive sequences. Because the DNA or RNA has been randomly sheared during library preparation, captured fragments are overlapping and unique.
Hybridization capture works well for genotyping and rare-variant detection. In contrast to amplicon sequencing where PCR primers must be designed to avoid any mismatched bases due to single nucleotide variations (SNVs) or indels, capture with hybridization probes is likely to perform better with respect to sequence complexity. Hybridization capture’s capacity for mutation discovery makes it particularly suited to cancer research. Since it can be designed for sequence complexity and scalability, this methodology is a better choice for exome sequencing, too.
The IDT xGen hybridization capture products includes a variety of predesigned panels and custom panels available in a range of panel sizes. An updated, automation-friendly protocol is available for high-throughput applications.
The xGen
Hyb Panel Design Tool can help guide you through the process of designing a panel specific for your research. Once your design is ready, simply order your panels and begin your research. The panels you design can also be queried or viewed at
any time.
Figure 1. Desired prepared library fragments are separated from off-target fragments using hybridization capture. First, xGen Universal Blockers are mixed with prepared library fragments DNA to prevent adapter to adapter hybridization. Blocked library fragments are then annealed to the 5’ biotinylated oligonucleotide probes from an xGen Predesigned Panel or an xGen Custom Hyb Panel. The probe and fragment duplexes are then separated from the unbound fragments by streptavidin-coated magnetic bead purification. The resulting library contains just the DNA or RNA fragments of interest.
Ordering
Are you looking for guidance on product compatibility or how to select the appropriate predesigned hybridization capture panel? Investigate the options and see our recommended product groupings with our xGen NGS Solutions Builder Tool.
xGen NGS Solutions Builder Tool
Let us help make selecting the right solutions easy!
Explore NGS options

 Processing
Processing